Difference between revisions of "More details on GSK-3"
(→Amino Acid Sequence) |
(→Structural details) |
||
| Line 15: | Line 15: | ||
==Structural details == | ==Structural details == | ||
| − | * GSK3 has the typical two-domain kinase fold with a beta-strand domain (residues 25−138) at the N-terminal end and an alpha-helical domain at the C-terminal end (residues 139−343). | + | * GSK3 has the typical two-domain kinase fold with a beta-strand domain (residues 25−138) at the N-terminal end and an alpha-helical domain at the C-terminal end (residues 139−343). <br> |
| − | * The ATP-binding site is at the interface of the alpha-helical and beta-strand domain and is bordered by the glycine-rich loop and the hinge. | + | * The ATP-binding site is at the interface of the alpha-helical and beta-strand domain and is bordered by the glycine-rich loop and the hinge. <br> |
| − | * The activation loop (residues 200−226) runs along the surface of the substrate binding groove. | + | * The activation loop (residues 200−226) runs along the surface of the substrate binding groove. <br> |
| − | * The C-terminal 39 residues (residues 344−382) are outside the core kinase fold and form a small domain that packs against the alpha-helical domain. | + | * The C-terminal 39 residues (residues 344−382) are outside the core kinase fold and form a small domain that packs against the alpha-helical domain. <br>[[image:gsk3_2.jpg|right|300 px]] |
| − | * The beta-strand domain consists of seven antiparallel beta-strands: strands 2−6 form a -barrel that is interrupted between strand 4 and 5 by a short helix (residue 96−102) that packs against the beta-barrel. | + | * The beta-strand domain consists of seven antiparallel beta-strands: strands 2−6 form a -barrel that is interrupted between strand 4 and 5 by a short helix (residue 96−102) that packs against the beta-barrel. <br> |
| − | * This helix is conserved in all kinases, and two of its residues play key roles in the catalytic activity of the enzyme. Arg 96 is involved in the alignment of the two domains. Glu 97 is positioned in the active site and forms a salt bridge with Lys 85, a key residue in catalysis. | + | * This helix is conserved in all kinases, and two of its residues play key roles in the catalytic activity of the enzyme. Arg 96 is involved in the alignment of the two domains. Glu 97 is positioned in the active site and forms a salt bridge with Lys 85, a key residue in catalysis.<br> |
| + | * Molecular weight: 46744.3 <br> | ||
| + | * Theoretical pI: 8.98 <br> | ||
| + | * Total number of negatively charged residues (Asp + Glu): 41 <br> | ||
| + | * Total number of positively charged residues (Arg + Lys): 50 <br> | ||
| − | [[image: | + | '''Atomic composition:''' |
| + | |||
| + | ** Carbon C 2085 <br> | ||
| + | ** Hydrogen H 3285 <br> | ||
| + | ** Nitrogen N 575 <br> | ||
| + | ** Oxygen O 618 <br> | ||
| + | ** Sulfur S 14 <br> | ||
| + | * Formula: C2085H3285N575O618S14 | ||
| + | * Total number of atoms: 6577 | ||
| + | |||
| + | '''Prediction search done on NetPhos 2.0 server for GSK3''' | ||
| + | |||
| + | Prediction search done on NetPhos 2.0 server, which produces neural network predictions for serine, threonine and tyrosine phosphorylation sites in eukaryotic proteins. | ||
| + | |||
| + | [[image:predication-gsk3.jpg|center|300 px]] | ||
==Amino Acid Sequence== | ==Amino Acid Sequence== | ||
Revision as of 04:08, 14 July 2006
Overview
Glycogen synthase kinase-3 (GSK-3) has recently emerged, in the field of medicinal chemistry, as one of the most attractive therapeutic targets for the development of selective inhibitors as promising new drugs for numerous serious pathologies, including Alzheimer's disease, stroke, bipolar disorders, chronic inflammatory processes, cancer, alopecia and Type II diabetes. The full potential of GSK-3 inhibitors is yet to be realised and the number of drug candidates being developed by both academic centres and pharmaceutical companies has increased exponentially in the last three years. This review discloses recent discoveries on peptides and small molecules targeting GSK-3. Antisense therapy for the modulation of GSK-3 expression is also discussed. Focusing attention on this exciting target could thus reap considerable clinical and economic rewards.
Protein name:Glycogen synthase kinase-3 alpha
Synonyms: EC 2.7.11.26; GSK-3 alpha
Gene name :Name: GSK3A
From : Homo sapiens (Human) [TaxID: 9606]
Function: Participates in the Wnt signaling pathway. Implicated in the hormonal control of several regulatory proteins including glycogen synthase, MYB and the transcription factor JUN. Phosphorylates JUN at sites proximal to its DNA-binding domain, thereby reducing its affinity for DNA.
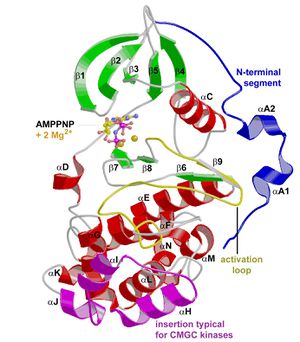
Structural details
- GSK3 has the typical two-domain kinase fold with a beta-strand domain (residues 25−138) at the N-terminal end and an alpha-helical domain at the C-terminal end (residues 139−343).
- The ATP-binding site is at the interface of the alpha-helical and beta-strand domain and is bordered by the glycine-rich loop and the hinge.
- The activation loop (residues 200−226) runs along the surface of the substrate binding groove.
- The C-terminal 39 residues (residues 344−382) are outside the core kinase fold and form a small domain that packs against the alpha-helical domain.
- The beta-strand domain consists of seven antiparallel beta-strands: strands 2−6 form a -barrel that is interrupted between strand 4 and 5 by a short helix (residue 96−102) that packs against the beta-barrel.
- This helix is conserved in all kinases, and two of its residues play key roles in the catalytic activity of the enzyme. Arg 96 is involved in the alignment of the two domains. Glu 97 is positioned in the active site and forms a salt bridge with Lys 85, a key residue in catalysis.
- Molecular weight: 46744.3
- Theoretical pI: 8.98
- Total number of negatively charged residues (Asp + Glu): 41
- Total number of positively charged residues (Arg + Lys): 50
Atomic composition:
- Carbon C 2085
- Hydrogen H 3285
- Nitrogen N 575
- Oxygen O 618
- Sulfur S 14
- Carbon C 2085
- Formula: C2085H3285N575O618S14
- Total number of atoms: 6577
Prediction search done on NetPhos 2.0 server for GSK3
Prediction search done on NetPhos 2.0 server, which produces neural network predictions for serine, threonine and tyrosine phosphorylation sites in eukaryotic proteins.
Amino Acid Sequence
GSK3B_HUMAN consists of 420 amino acids sequemnce.