Difference between revisions of "Hormone Sensitive Lipase"
From DolceraWiki
| (8 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| + | ==Overview== | ||
| + | ===Hormone Sensitive Lipase (HSL)=== | ||
| + | * Hormone-sensitive lipase is an enzyme expressed in multiple tissues. It plays number of roles in lipid metabolism. | ||
| + | * Major isoform of HSL gene is a single polypeptide with molecular mass of approximately 84kDa, its has 3 major domains catalytic, regulatory domain congaing several plosphorylation sistes, and n-terminal domain involved in protein-protein and protein-lipid interactions. | ||
| + | * Altererd expression of HSL in different celltypes may be associated with several pathological states, includng obesity, atherosclerosis, Type II Diabetes. | ||
| + | * HSL has lipolytic activity against not only triacylglycerol, but also against diacylglycerol, monoacylglycerol, and cholesteryl esters. | ||
| + | * Gene and protein structure of the HSL given below. | ||
| + | [[Image:Gene_Map.jpg|thumb|center|750px|Gene: exon/intron organization of the human HSL gene. Exons T and A encode 300 and 43 additional amino acids respectively, located at the N-terminus.<BR> | ||
| + | Protein: Linear representation of the amino acid sequence of adipocyte HSL (numbering of the rat sequence).<BR> | ||
| + | Domains: N-terminal binding domain, a C-terminal catalytic domain, harbouring the catalytic triad and a regulatory module containing multiple phosphorylation sites. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=DisplayFiltered&DB=pubmed Source]]] | ||
| + | * Fates of HSL products in different cell types. | ||
| + | [[Image:HSL_products.jpg|center|Fates of HSL products]] | ||
| + | * [[HSL protein sequence]] - [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&qty=1&c_start=1&list_uids=145559491&uids=&dopt=fasta&dispmax=5&sendto=&from=begin&to=end&extrafeatpresent=1&ef_CDD=8&ef_MGC=16&ef_HPRD=32&ef_STS=64&ef_tRNA=128&ef_microRNA=256 Source1], [http://www.expasy.org/uniprot/Q05469 Source2] | ||
| + | * [[HSL mRNA sequence]] - [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=NM_005357.2 Source] | ||
| + | |||
| + | ===HSL, a drug target for Diabetes and Metabolic Disease=== | ||
| + | A few points suggesting HSL as a drug tareget for Type 2 Diabetes and Metabolic Disease: | ||
| + | * There are conflicting data on whether altered expression or activity of HSL is involved in familial combined hyperlipidaemia. Similar debate surrounds the role of altered levels of HSL in obesity and in insulin resistance, two linked conditions which predispose to Type II Diabetes. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed Source] | ||
| + | * Elevated levels of plasma nonesterified fatty acids are associated with insulin resistance. | ||
| + | * A promoter variant of HSL,−60C→G, has been implicated in a number of conditions, which exhibits a 40% decrease in promoter activity, has been associated with increased insulin sensitivity in women and decreased levels of plasma non-esterified fatty acid in men. | ||
| + | * Population studies suggest that a polymorphic marker in the HSL gene is in linkage disequilibrium with an allele that increases susceptibility to abdominal obesity, which is itself a risk factor for Type II diabetes. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=9867220&query_hl=12&itool=pubmed_docsum Source] | ||
| + | * Over expression of HSL prevents lipid accumulation (from cell line studies). [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=7769105&query_hl=14&itool=pubmed_docsum Source] | ||
| + | |||
| + | ===Type 2 Diabetes=== | ||
| + | * Type 2 Diabetes Mellitus is a group of metabolic diseases characterized by high blood sugar (glucose) levels, which result from defects in insulin secretion, or insulin resistance, or both. | ||
| + | * Insulin, a hormone produced by the pancreas, which controls blood glucose levels. | ||
| + | * Insulin triggers glucose uptake by the cells. Some part of the glucose can be converted to concentrated energy sources like glycogen, triacylglycerols, or fatty acids. | ||
| + | |||
| + | ===Metabolic Disease=== | ||
| + | * A person with abnormal glucose tolerance (IGT or diabetes) will be found to have at least one or more of the other cardiovascular disease (CVD) risk components. | ||
| + | * This clustering has been labelled variously as Syndrome X, the Insulin Resistance Syndrome, the Metabolic Syndrome or Metabolic disease. | ||
| + | |||
| + | ==Non-patent Literature Search and Analysis== | ||
| + | |||
| + | * In non-patent literature searching we got 17 records/articles. Out of these 8 articles are talking about chemical compounds that are involved in inhibition or stimulation of hormone-sensitive lipase (HSL) and 9 articles are scientific review papers. | ||
| + | |||
| + | ====Search Strategy==== | ||
| + | {|border="1" cellspacing="0" cellpadding="4" width="100%" | ||
| + | |align = "center" bgcolor = "#FFFF99" rowspan = "2"|'''S.No.''' | ||
| + | |align = "center" bgcolor = "#FFFF99" rowspan = "2"|'''Database''' | ||
| + | |align = "center" bgcolor = "#FFFF99" colspan = "2"|'''Key-Words''' | ||
| + | |align = "center" bgcolor = "#FFFF99" rowspan = "2"|'''Search Queries''' | ||
| + | |align = "center" bgcolor = "#FFFF99" rowspan = "2"|'''No. of Records''' | ||
| + | |- | ||
| + | |align = "center" bgcolor = "#FFFF99"|'''Main Terms''' | ||
| + | |align = "center" bgcolor = "#FFFF99"|'''Alternative Terms''' | ||
| + | |- | ||
| + | |1 | ||
| + | |BIOSIS | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L4 1576 (HORMONE (W) SENSITIVE (2A) LIPASE#) OR HSL OR LHS OR LIPE OR (HORMONESENSITIVE (A) LIPASE#) | ||
| + | * L5 266641 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L6 30 L4 (S) L5 | ||
| + | |30 | ||
| + | |- | ||
| + | |2 | ||
| + | |EMBASE | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS, L11706, U40001, Q05469 | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L2 1203 (HORMONE (W) SENSITIVE (2A) LIPASE# ) OR (HORMONESENSITIVE (W) LIPASE# ) OR LIPE OR HSL OR LHS OR L11706 OR U40001 OR Q05469 | ||
| + | * L3 258095 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L4 32 L2 (S) L3 | ||
| + | | 32 | ||
| + | |- | ||
| + | |3 | ||
| + | |DISSABS | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L1 131 (HORMONE (W) SENSITIVE (2A) LIPASE ) OR (HORMONESENSITIVE (W) LIPASE ) OR LIPE OR HSL OR LHS | ||
| + | * L2 6000 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L3 5 L1 (S) L2 | ||
| + | |5 | ||
| + | |- | ||
| + | |4 | ||
| + | |MEDLINE | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS, L11706, U40001, Q05469 | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L1 1331 (HORMONE (W) SENSITIVE (2A) LIPASE ) OR (HORMONESENSITIVE (W) LIPASE ) OR LIPE OR HSL OR LHS OR L11706 OR U40001 OR Q05469 | ||
| + | * L2 286108 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L3 29 L1 (S) L2 | ||
| + | | 29 | ||
| + | |- | ||
| + | |5 | ||
| + | |BIOTECHNO | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS, L11706, U40001 | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L1 398 (HORMONE (W) SENSITIVE (2A) LIPASE#) OR (HORMONESENSITIVE (W) LIPASE#) OR LIPE OR HSL OR LHS OR L11706 OR U40001 | ||
| + | * L2 18139 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L3 10 L1 (S) L2 | ||
| + | | 10 | ||
| + | |- | ||
| + | |6 | ||
| + | |BIOTECHABS | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS, L11706, U40001, Q05469 | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L1 56 (HORMONE (W) SENSITIVE (2A) LIPASE#) OR (HORMONESENSITIVE (W) LIPASE#) OR LIPE OR HSL OR LHS | ||
| + | * L2 9716 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L3 5 L1 (S) L2 | ||
| + | | 5 | ||
| + | |- | ||
| + | |7 | ||
| + | |CONFSCI | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L1 90 (HORMONE (W) SENSITIVE (2A) LIPASE#) OR (HORMONESENSITIVE (W) LIPASE#) OR LIPE OR HSL OR LHS | ||
| + | * L2 1257 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L3 0 L1 (S) L2 | ||
| + | | 0 | ||
| + | |- | ||
| + | |8 | ||
| + | |NTIS | ||
| + | | | ||
| + | * Hormone sensitive lipase | ||
| + | * Type 2 diabetes | ||
| + | | | ||
| + | * Hormone-sensitive lipase, Lipase-hormone sensitive, Hormonesensitive lipase, LIPE, HSL, LHS | ||
| + | * Diabetes, NIDDM, IDDM, T2DM | ||
| + | | | ||
| + | * L1 90 (HORMONE (W) SENSITIVE (2A) LIPASE#) OR (HORMONESENSITIVE (W) LIPASE#) OR LIPE OR HSL OR LHS | ||
| + | * L2 1257 DIABET? OR NIDDM OR IDDM OR T2DM | ||
| + | * L3 0 L1 (S) L2 | ||
| + | | 0 | ||
| + | |- | ||
| + | |colspan="6"| | ||
| + | * Total number of records = 106 | ||
| + | * After removing duplicates number of records = 49 | ||
| + | * Total on-targets (after removing duplicates) = 17 | ||
| + | * Accuracy of the query = 35% | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ==== Scientific papers on chemical compounds inhibiting or stimulating HSL ==== | ||
| + | * Following is the distribution of the 8 articles based on Function of the compounds. | ||
| + | * Numbers in brackets indicate number of records in the concerned topic. | ||
| + | * Highlighted (in red color) numbers refer serial number in the [[Analysis Table| analysis table]]. | ||
| + | [[Image:non_ip_1.jpg|750px|thumb|center|Distribution of non-patent articles]] | ||
| + | |||
| + | * Detailed analysis of the above records is available at the following link: [[Analysis Table]] | ||
| + | the | ||
| + | |||
| + | ==== General scientific studies on HSL found from STN==== | ||
| + | * Off the 17 articles, 9 articles are talking about variations in the HSL gene and review papers. Variations discussed are C-60G polymorphism in the promoter of the HSL gene and a dinucleotide repeat in the gene, which are associated with Type 2 diabetes, suggesting the putative role of the HSL in Type 2 diabetes. Other articles are reviews saying HSL is a potential target for treating or curing Type 2 diabetes. | ||
| + | * The following hyperlink takes us to the above mentioned 9 articles: [[Scientific studies of HSL]] | ||
| + | |||
| + | ==Patentability Search== | ||
| + | ===Sequence Search and Analysis=== | ||
| + | * Sequence search on NCBI. | ||
| + | ** Got 16 relevant patents. | ||
| + | ** [[Media:NCBI_spreadsheet_2.xls|Spread sheet]] | ||
| + | * Sequence search on DGENE. | ||
| + | ** Got 42 relevant patents. | ||
| + | ** [[Media:DGENE_spreadsheet_2.xls|Spread sheet]] | ||
| + | * Sequence search on PCTGEN. | ||
| + | ** Got 16 relevant patents. | ||
| + | ** [[Media:PCTGEN_spreadsheet_2.xls|Spread sheet]] | ||
| + | |||
| + | ===Sequence Search Stratagy=== | ||
| + | |||
| + | {|border="1" cellspacing="0" cellpadding="4" width="100%" | ||
| + | |'''S.No.''' | ||
| + | |'''Database''' | ||
| + | |'''Sequence Query''' | ||
| + | |'''Key Words '''(to restrict unrelated patents) | ||
| + | |'''Hits''' | ||
| + | |- | ||
| + | |1 | ||
| + | |NCBI (Blast) | ||
| + | | | ||
| + | * HSL protein | ||
| + | | | ||
| + | |||
| + | |13 | ||
| + | |- | ||
| + | |2 | ||
| + | |NCBI (Blast) | ||
| + | | | ||
| + | * HSL mRNA | ||
| + | | | ||
| + | |||
| + | |19 | ||
| + | |- | ||
| + | |3 | ||
| + | |DGENE (Blast search) | ||
| + | | | ||
| + | * HSL protein | ||
| + | * HSL mRNA | ||
| + | | | ||
| + | * diabetes | ||
| + | * NIDDM | ||
| + | * IDDM | ||
| + | * T2DM | ||
| + | |61 | ||
| + | |- | ||
| + | |- | ||
| + | |4 | ||
| + | |PCTGEN (Blast search) | ||
| + | | | ||
| + | * HSL protein | ||
| + | * HSL mRNA | ||
| + | | | ||
| + | |83 | ||
| + | |- | ||
| + | |colspan="5"| | ||
| + | * '''In NCBI got 16 relevat patents.''' | ||
| + | * '''In DGENE got 42 relevant patents.''' | ||
| + | * '''In PCTGEN got 16 relevant patents.''' | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ==<span style="color:#C41E3A">Like this report?</span>== | ||
| + | <p align="center"> '''This is only a sample report with brief analysis''' <br> | ||
| + | '''Dolcera can provide a comprehensive report customized to your needs'''</p> | ||
| + | {|border="2" cellspacing="0" cellpadding="4" align="center" " | ||
| + | |style="background:lightgrey" align = "center" colspan = "3"|'''[mailto:info@dolcera.com <span style="color:#0047AB">Buy the customized report from Dolcera</span>]''' | ||
| + | |- | ||
| + | | align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services Patent Analytics Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/business-research-services Market Research Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/tools/patent-dashboard Purchase Patent Dashboard] | ||
| + | |- | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services/patent-search/patent-landscapes Patent Landscape Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/research-processes Dolcera Processes] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/industries Industry Focus] | ||
| + | |- | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services/patent-search/patent-landscapes Patent Search Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services/alerts-and-updates Patent Alerting Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/tools Dolcera Tools] | ||
| + | |- | ||
| + | |} | ||
| + | <br> | ||
| + | |||
| + | ===Micropatent Search and Analysis=== | ||
| + | * The following is the link to search strategy on Micropat [[Search on micropat]] | ||
| + | * The following provides an example of how patents were analyzed[[Sample patent analysis]] | ||
| + | |||
| + | * From Micropat search, 769 patents were analyzed out of which 41 are on target. | ||
| + | |||
| + | ==== Final Patent list (with bibliographic information) ==== | ||
| + | * After removing duplicates and family members of relevant patents found from NCBI, PCT Gene, DGENE and Micropat, we got 96 relevant patents. | ||
| + | * [[Media:finla_list_96_1.xls|Final list of patents]] | ||
| + | |||
| + | ==== Overall IP activity==== | ||
| + | * The below graph indicates the overall patenting trend for the 96 relevant patents over time | ||
| + | |||
| + | [[Image:BMS_IP_A_1.jpg|Overall IP Activity|thumb|450px|center]] | ||
| + | |||
| + | ====Key Players (those with more than 2 patents)==== | ||
| + | * Key players in the field (total 66 records) are Novo Nordisk, Sanofi Aventis, Incyte, Curagen, Hyseq, Alizyme Therapeutics, Genentech, Human Genome Sciences, Takeda, Alteon, Bristol-Myers Squibb, Eli Lilly ,Genset, ISIS Pharmaceuticals, Japan Tobacco, Novartis. | ||
| + | [[Image:BMS_A_I.jpg|600px|center|Key Players|thumb]] | ||
| + | |||
| + | ==== Other players (those with 1 patent) ==== | ||
| + | * In all, other players have 30 patents to their name. | ||
| + | {|border="1" cellspacing="0" cellpadding="4" width="60%" | ||
| + | |'''Assignee''' | ||
| + | |'''No. of Patents/Publications''' | ||
| + | |- | ||
| + | |Arena Pharmaceuticals | ||
| + | |1 | ||
| + | |- | ||
| + | |Ares Trading | ||
| + | |1 | ||
| + | |- | ||
| + | |Asahi Kasei Pharma Corporation | ||
| + | |1 | ||
| + | |- | ||
| + | |Bionomics | ||
| + | |1 | ||
| + | |- | ||
| + | |Compugen | ||
| + | |1 | ||
| + | |- | ||
| + | |Epiontis | ||
| + | |1 | ||
| + | |- | ||
| + | |Euroscreen | ||
| + | |1 | ||
| + | |- | ||
| + | |Exelixis | ||
| + | |1 | ||
| + | |- | ||
| + | |F. Hoffmann La Roche Ag | ||
| + | |1 | ||
| + | |- | ||
| + | |Five Prime Therapeutics | ||
| + | |1 | ||
| + | |- | ||
| + | |Forschungsverbund Berlin | ||
| + | |1 | ||
| + | |- | ||
| + | |Genetics Iinstitute LLC, Ono Pharmaceuticals | ||
| + | |1 | ||
| + | |- | ||
| + | |GPC Biotech | ||
| + | |1 | ||
| + | |- | ||
| + | |Immunex | ||
| + | |1 | ||
| + | |- | ||
| + | |Linkagene Ltd. | ||
| + | |1 | ||
| + | |- | ||
| + | |Merck & Co | ||
| + | |1 | ||
| + | |- | ||
| + | |Metabolex | ||
| + | |1 | ||
| + | |- | ||
| + | |Micromet AG | ||
| + | |1 | ||
| + | |- | ||
| + | |Nuvelo | ||
| + | |1 | ||
| + | |- | ||
| + | |Ottawa Health Research Institute | ||
| + | |1 | ||
| + | |- | ||
| + | |President & Fellows Of Harvard College | ||
| + | |1 | ||
| + | |- | ||
| + | |Protein Design Labs | ||
| + | |1 | ||
| + | |- | ||
| + | |Regents Of The University Of California | ||
| + | |1 | ||
| + | |- | ||
| + | |Rutgers,The State University Of New Jersey | ||
| + | |1 | ||
| + | |- | ||
| + | |Schering Corporation | ||
| + | |1 | ||
| + | |- | ||
| + | |Sequenom | ||
| + | |1 | ||
| + | |- | ||
| + | |The Buck Institute For Age Research | ||
| + | |1 | ||
| + | |- | ||
| + | |The Regents Of The University Of Michigan | ||
| + | |1 | ||
| + | |- | ||
| + | |University Of Maryland Baltimore | ||
| + | |1 | ||
| + | |- | ||
| + | |Yissum Research Development Company Of The Hebrew University Of Jerusalem | ||
| + | |1 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ==Patent Analysis== | ||
| + | ====Patent Distribution (Sample analysis of 25 records only) ==== | ||
| + | * HSL refers to hormone-sensitive lipase. | ||
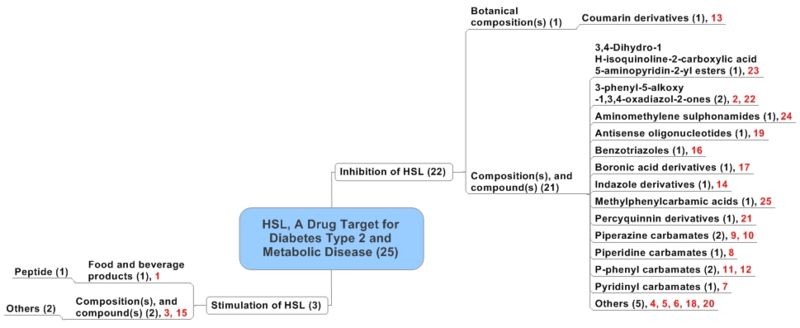
| + | * Patents distributed based on patent focus and mechanism of the action of the compounds. | ||
| + | * The number of patents referring to a class of compounds is given in the brackets in the figure. | ||
| + | * The below map is a sample representation. | ||
| + | [[Image:BMS_P_D.jpg|Patent Distribution|800px|center|thumb]] | ||
| + | * Details about patents available from the following hyperlink, [[Media:BMS_Spread_Sheet_210607_2.xls|analysis sheet]] | ||
| + | |||
| + | ==Dashboard (Sample based on 25 analyzed records) == | ||
| + | * '''[http://client.dolcera.com/dashboard/dashboard.html?workfile_id=22 Dashboard on patents and analysis]''' | ||
| + | * The given dashboard is a sample representation. | ||
| + | |||
| + | ==Interactive Mindmap== | ||
| + | <mm>[[Mmap22.mm]]</mm> | ||
| + | |||
| + | ==<span style="color:#C41E3A">Like this report?</span>== | ||
| + | <p align="center"> '''This is only a sample report with brief analysis''' <br> | ||
| + | '''Dolcera can provide a comprehensive report customized to your needs'''</p> | ||
| + | {|border="2" cellspacing="0" cellpadding="4" align="center" " | ||
| + | |style="background:lightgrey" align = "center" colspan = "3"|'''[mailto:info@dolcera.com <span style="color:#0047AB">Buy the customized report from Dolcera</span>]''' | ||
| + | |- | ||
| + | | align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services Patent Analytics Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/business-research-services Market Research Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/tools/patent-dashboard Purchase Patent Dashboard] | ||
| + | |- | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services/patent-search/patent-landscapes Patent Landscape Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/research-processes Dolcera Processes] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/industries Industry Focus] | ||
| + | |- | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services/patent-search/patent-landscapes Patent Search Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/services/ip-patent-analytics-services/alerts-and-updates Patent Alerting Services] | ||
| + | |align = "center"| [http://www.dolcera.com/website_prod/tools Dolcera Tools] | ||
| + | |- | ||
| + | |} | ||
| + | <br> | ||
| + | |||
| + | ==Contact Dolcera== | ||
| + | |||
| + | {| style="border:1px solid #AAA; background:#E9E9E9" align="center" | ||
| + | |- | ||
| + | ! style="background:lightgrey" | Samir Raiyani | ||
| + | |- | ||
| + | | '''Email''': [mailto:info@dolcera.com info@dolcera.com] | ||
| + | |- | ||
| + | | '''Phone''': +1-650-269-7952 | ||
| + | |} | ||
Latest revision as of 05:11, 21 September 2010
Contents
Overview
Hormone Sensitive Lipase (HSL)
- Hormone-sensitive lipase is an enzyme expressed in multiple tissues. It plays number of roles in lipid metabolism.
- Major isoform of HSL gene is a single polypeptide with molecular mass of approximately 84kDa, its has 3 major domains catalytic, regulatory domain congaing several plosphorylation sistes, and n-terminal domain involved in protein-protein and protein-lipid interactions.
- Altererd expression of HSL in different celltypes may be associated with several pathological states, includng obesity, atherosclerosis, Type II Diabetes.
- HSL has lipolytic activity against not only triacylglycerol, but also against diacylglycerol, monoacylglycerol, and cholesteryl esters.
- Gene and protein structure of the HSL given below.
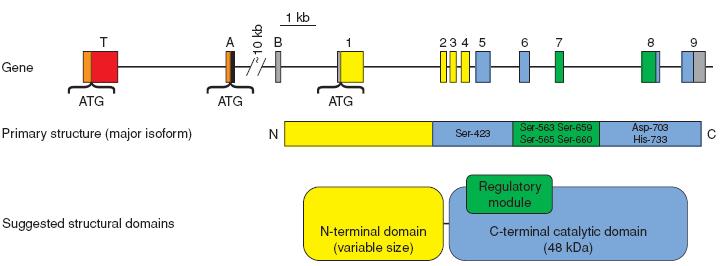
Gene: exon/intron organization of the human HSL gene. Exons T and A encode 300 and 43 additional amino acids respectively, located at the N-terminus.
Protein: Linear representation of the amino acid sequence of adipocyte HSL (numbering of the rat sequence).
Domains: N-terminal binding domain, a C-terminal catalytic domain, harbouring the catalytic triad and a regulatory module containing multiple phosphorylation sites. Source
Protein: Linear representation of the amino acid sequence of adipocyte HSL (numbering of the rat sequence).
Domains: N-terminal binding domain, a C-terminal catalytic domain, harbouring the catalytic triad and a regulatory module containing multiple phosphorylation sites. Source
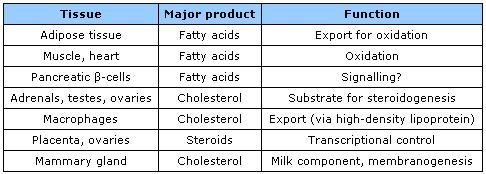
- Fates of HSL products in different cell types.
HSL, a drug target for Diabetes and Metabolic Disease
A few points suggesting HSL as a drug tareget for Type 2 Diabetes and Metabolic Disease:
- There are conflicting data on whether altered expression or activity of HSL is involved in familial combined hyperlipidaemia. Similar debate surrounds the role of altered levels of HSL in obesity and in insulin resistance, two linked conditions which predispose to Type II Diabetes. Source
- Elevated levels of plasma nonesterified fatty acids are associated with insulin resistance.
- A promoter variant of HSL,−60C→G, has been implicated in a number of conditions, which exhibits a 40% decrease in promoter activity, has been associated with increased insulin sensitivity in women and decreased levels of plasma non-esterified fatty acid in men.
- Population studies suggest that a polymorphic marker in the HSL gene is in linkage disequilibrium with an allele that increases susceptibility to abdominal obesity, which is itself a risk factor for Type II diabetes. Source
- Over expression of HSL prevents lipid accumulation (from cell line studies). Source
Type 2 Diabetes
- Type 2 Diabetes Mellitus is a group of metabolic diseases characterized by high blood sugar (glucose) levels, which result from defects in insulin secretion, or insulin resistance, or both.
- Insulin, a hormone produced by the pancreas, which controls blood glucose levels.
- Insulin triggers glucose uptake by the cells. Some part of the glucose can be converted to concentrated energy sources like glycogen, triacylglycerols, or fatty acids.
Metabolic Disease
- A person with abnormal glucose tolerance (IGT or diabetes) will be found to have at least one or more of the other cardiovascular disease (CVD) risk components.
- This clustering has been labelled variously as Syndrome X, the Insulin Resistance Syndrome, the Metabolic Syndrome or Metabolic disease.
Non-patent Literature Search and Analysis
- In non-patent literature searching we got 17 records/articles. Out of these 8 articles are talking about chemical compounds that are involved in inhibition or stimulation of hormone-sensitive lipase (HSL) and 9 articles are scientific review papers.
Search Strategy
| S.No. | Database | Key-Words | Search Queries | No. of Records | |
| Main Terms | Alternative Terms | ||||
| 1 | BIOSIS |
|
|
|
30 |
| 2 | EMBASE |
|
|
|
32 |
| 3 | DISSABS |
|
|
|
5 |
| 4 | MEDLINE |
|
|
|
29 |
| 5 | BIOTECHNO |
|
|
|
10 |
| 6 | BIOTECHABS |
|
|
|
5 |
| 7 | CONFSCI |
|
|
|
0 |
| 8 | NTIS |
|
|
|
0 |
| |||||
Scientific papers on chemical compounds inhibiting or stimulating HSL
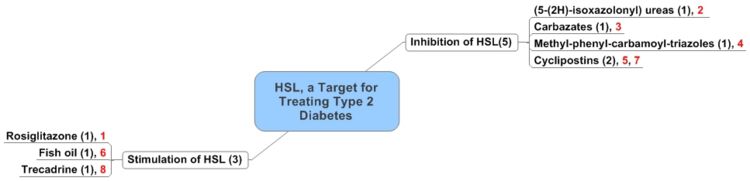
- Following is the distribution of the 8 articles based on Function of the compounds.
- Numbers in brackets indicate number of records in the concerned topic.
- Highlighted (in red color) numbers refer serial number in the analysis table.
- Detailed analysis of the above records is available at the following link: Analysis Table
the
General scientific studies on HSL found from STN
- Off the 17 articles, 9 articles are talking about variations in the HSL gene and review papers. Variations discussed are C-60G polymorphism in the promoter of the HSL gene and a dinucleotide repeat in the gene, which are associated with Type 2 diabetes, suggesting the putative role of the HSL in Type 2 diabetes. Other articles are reviews saying HSL is a potential target for treating or curing Type 2 diabetes.
- The following hyperlink takes us to the above mentioned 9 articles: Scientific studies of HSL
Patentability Search
Sequence Search and Analysis
- Sequence search on NCBI.
- Got 16 relevant patents.
- Spread sheet
- Sequence search on DGENE.
- Got 42 relevant patents.
- Spread sheet
- Sequence search on PCTGEN.
- Got 16 relevant patents.
- Spread sheet
Sequence Search Stratagy
| S.No. | Database | Sequence Query | Key Words (to restrict unrelated patents) | Hits |
| 1 | NCBI (Blast) |
|
13 | |
| 2 | NCBI (Blast) |
|
19 | |
| 3 | DGENE (Blast search) |
|
|
61 |
| 4 | PCTGEN (Blast search) |
|
83 | |
| ||||
Like this report?
This is only a sample report with brief analysis
Dolcera can provide a comprehensive report customized to your needs
Micropatent Search and Analysis
- The following is the link to search strategy on Micropat Search on micropat
- The following provides an example of how patents were analyzedSample patent analysis
- From Micropat search, 769 patents were analyzed out of which 41 are on target.
Final Patent list (with bibliographic information)
- After removing duplicates and family members of relevant patents found from NCBI, PCT Gene, DGENE and Micropat, we got 96 relevant patents.
- Final list of patents
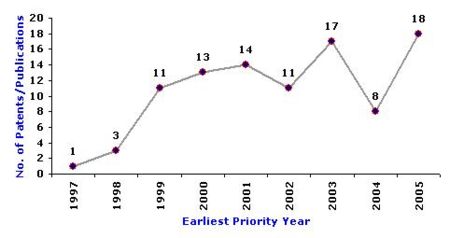
Overall IP activity
- The below graph indicates the overall patenting trend for the 96 relevant patents over time
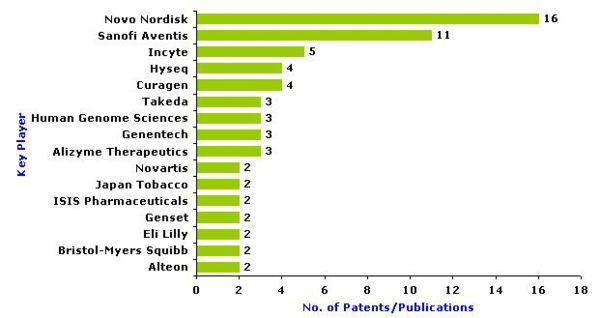
Key Players (those with more than 2 patents)
- Key players in the field (total 66 records) are Novo Nordisk, Sanofi Aventis, Incyte, Curagen, Hyseq, Alizyme Therapeutics, Genentech, Human Genome Sciences, Takeda, Alteon, Bristol-Myers Squibb, Eli Lilly ,Genset, ISIS Pharmaceuticals, Japan Tobacco, Novartis.
Other players (those with 1 patent)
- In all, other players have 30 patents to their name.
| Assignee | No. of Patents/Publications |
| Arena Pharmaceuticals | 1 |
| Ares Trading | 1 |
| Asahi Kasei Pharma Corporation | 1 |
| Bionomics | 1 |
| Compugen | 1 |
| Epiontis | 1 |
| Euroscreen | 1 |
| Exelixis | 1 |
| F. Hoffmann La Roche Ag | 1 |
| Five Prime Therapeutics | 1 |
| Forschungsverbund Berlin | 1 |
| Genetics Iinstitute LLC, Ono Pharmaceuticals | 1 |
| GPC Biotech | 1 |
| Immunex | 1 |
| Linkagene Ltd. | 1 |
| Merck & Co | 1 |
| Metabolex | 1 |
| Micromet AG | 1 |
| Nuvelo | 1 |
| Ottawa Health Research Institute | 1 |
| President & Fellows Of Harvard College | 1 |
| Protein Design Labs | 1 |
| Regents Of The University Of California | 1 |
| Rutgers,The State University Of New Jersey | 1 |
| Schering Corporation | 1 |
| Sequenom | 1 |
| The Buck Institute For Age Research | 1 |
| The Regents Of The University Of Michigan | 1 |
| University Of Maryland Baltimore | 1 |
| Yissum Research Development Company Of The Hebrew University Of Jerusalem | 1 |
Patent Analysis
Patent Distribution (Sample analysis of 25 records only)
- HSL refers to hormone-sensitive lipase.
- Patents distributed based on patent focus and mechanism of the action of the compounds.
- The number of patents referring to a class of compounds is given in the brackets in the figure.
- The below map is a sample representation.
- Details about patents available from the following hyperlink, analysis sheet
Dashboard (Sample based on 25 analyzed records)
- Dashboard on patents and analysis
- The given dashboard is a sample representation.
Interactive Mindmap
Like this report?
This is only a sample report with brief analysis
Dolcera can provide a comprehensive report customized to your needs
Contact Dolcera
| Samir Raiyani |
|---|
| Email: info@dolcera.com |
| Phone: +1-650-269-7952 |