Difference between revisions of "RNAi Database"
From DolceraWiki
(→Effector RNA molecules) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 7: | Line 7: | ||
RNAi pathways are guided by small RNAs that include | RNAi pathways are guided by small RNAs that include | ||
| − | ''' | + | '''siRNA:''' |
* Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of 20-25 nucleotide-long double-stranded RNA molecules. | * Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of 20-25 nucleotide-long double-stranded RNA molecules. | ||
* SiRNAs can also be exogenously (artificially) introduced into cells by various transfection methods to bring about the specific knockdown of a gene of interest. [http://en.wikipedia.org/wiki/SiRNA Source] | * SiRNAs can also be exogenously (artificially) introduced into cells by various transfection methods to bring about the specific knockdown of a gene of interest. [http://en.wikipedia.org/wiki/SiRNA Source] | ||
| Line 27: | Line 27: | ||
==Cancer specific Targets == | ==Cancer specific Targets == | ||
| − | [[Image:Targets.jpg|thumb|center| | + | [[Image:Targets.jpg|thumb|center|800 px|'''Cancer specific Targets'''[http://journals.prous.com/journals/servlet/xmlxsl/pk_journals.xml_summaryn_pr?p_JournalId=3&p_RefId=985937 Source]]] |
==Search strategy== | ==Search strategy== | ||
Latest revision as of 18:37, 14 July 2009
Contents
Overview
- RNA interference (RNAi), is a technique in which exogenous, double-stranded RNAs (dsRNAs) that are complimentary to known mRNAs, are introduced into a cell to specifically destroy that particular mRNA, thereby diminishing or abolishing gene expression.
- This technology considerably bolsters functional genomics to aid in the identification of novel genes involved in disease processes and thus can be used for medicament and for delivery as therapeutics. Source
- RNA interference was known by other names, including post transcriptional gene silencing and quelling. Source
Effector RNA molecules
RNAi pathways are guided by small RNAs that include
siRNA:
- Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of 20-25 nucleotide-long double-stranded RNA molecules.
- SiRNAs can also be exogenously (artificially) introduced into cells by various transfection methods to bring about the specific knockdown of a gene of interest. Source
miRNA:
- microRNAs (miRNA) are single-stranded RNA molecules of about 21–23 nucleotides in length, which regulate gene expression.
- miRNAs are encoded by genes from whose DNA they are transcribed but miRNAs are not translated into protein (non-coding RNA); instead each primary transcript (a pri-miRNA) is processed into a short stem-loop structure called a pre-miRNA and finally into a functional miRNA.
- Mature miRNA molecules are partially complementary to one or more messenger RNA (mRNA) molecules, and their main function is to down-regulate gene expression. Source
shRNA:
- A small hairpin RNA or short hairpin RNA (shRNA) is a sequence of RNA that makes a tight hairpin turn that can be used to silence gene expression via RNA interference.
shRNA uses a vector introduced into cells and utilizes the U6 promoter to ensure that the shRNA is always expressed. This vector is usually passed on to daughter cells, allowing the gene silencing to be inherited. The shRNA hairpin structure is cleaved by the cellular machinery into siRNA, which is then bound to the RNA-induced silencing complex (RISC). This complex binds to and cleaves mRNAs which match the siRNA that is bound to it. Source
Others:
- In addition to miRNAs and siRNAs, other innate RNAi effectors have been identified.
- One class of these is the Piwi-interacting RNAs (piRNAs). piRNAs seem to be uniquely expressed in the mammalian germline, particularly in the testes. The functional role of piRNAs is currently unclear, but a role in spermatogenesis is likely.
- A number of other small RNAs associated with RNAi have been identified in different species, including trans-activating siRNAs (tasiRNAs), studied in plants and nematodes, and small scan RNAs (ScnRNAs), found in Tetrahymena. Source
Cancer specific Targets
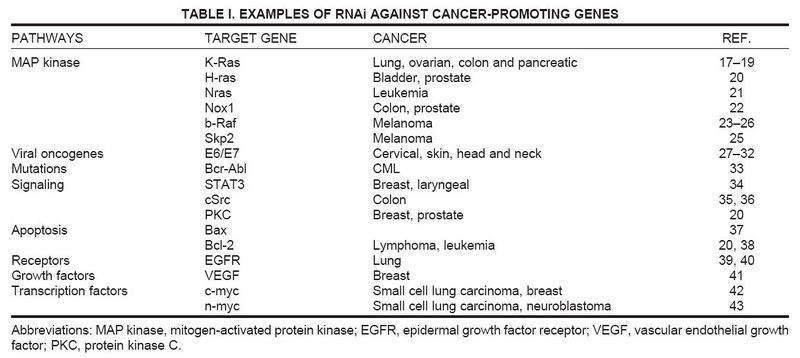
Cancer specific TargetsSource
Search strategy
| S.No | Concept | Search Scope | Query | No. of Hits |
| 1 | RNAi | Full Patent Spec | (siRNA*1 OR (short ADJ1 interfering ADJ1 RNA*1) OR (short ADJ1 interfering ADJ1 nucleic ADJ1 acid*1) OR (short ADJ1 interfering ADJ1 NA*1) OR siNA OR siNAs OR (si ADJ1 RNA*1) OR (Small adj1 interfering adj1 RNA*)OR miRNA or (microRNA* or micro adj1 RNA*)OR shRNA or (small adj1 hairpin adj1 RNA*)OR piRNA or (Piwi adj1 interacting adj1 RNA*)OR tasiRNAs or (trans adj1 activating adj1 siRNA*) OR ScnRNAs or (small adj1 scan adj1 RNA*)OR RNAi or(RNA adj1 interference)) | 28315 |
| 2 | GPX-1 | Full Patent Spec | (Glutathione NEAR2 peroxidase ADJ1 1) OR (Se ADJ GPx1)OR GSHPX1 OR MGC14399 OR MGC88245 OR GPX1 | 432 |
| 3 | RNAi and GPX-1 | 1 And 2 | 97 (Unique 58) | |
| 4 | c-MYC | Full Patent Spec | cmyc or (c adj1 myc) or MYC or "c-Myc" | 48132 |
| 5 | c-MYC and RNAi | 1 and 4 | 5811 | |
| 6 | IPC | Current IPC-R a61k or c12q or c12n or c07k or c07h | 1679356 | |
| 7 | c-MYC and RNAi + IPC | 5 and 6 | 5037 (Unique 2927) | |
| 8 | Bcl2 | Full Patent Spec | (Bcl2 OR (Bcell ADJ1 CLL) OR (Bcell ADJ1 Lymphoma2 ) OR DKFZp781P2092) and Current IPC-R a61k or c12q or c12n or c07k or c07h | 3837 |
| 9 | Bcl2 + RNAi +IPC | 1 and 8 | 1038 (Unique 562) | |
| 10 | K-RAS | Full Patent Spec | (k adj1 RaS) or (V adj1 Ki adj1 ras2 adj1 Kirsten adj1 rat adj1 sarcoma adj1 viral adj1 oncogene adj1 homolog) or (KRAS) or (C adj1 K adj1 RAs) or (K adj 1 RAS2A) or (K adj1 RAS2B) or (K adj1 RAS4A) or (K adj1 RAS4B) or (KI adj1 RAS) or KRAS1 or KRAS2 or NS3 or RASK2 or (K adj1 ras adj1 p21 adj1 protein) or (KI adj1 RAS) or (KRAS) or KRAS1 or KRAS2 or (Kirsten adj1 rat adj1 sarcoma adj1 2 adj1 viral adj1 oncogene adj1 homolog) or NS3 or (PR310 adj1 c adj1 K adj1 ras adj1 oncogene) or RASK2 or (c adj1 K adj1 ras2 adj1 protein) or (c adj1 Kirsten adj1 ras adj1 protein) or (cellular adj1 c adj1 Ki adj1 ras2 adj1 proto adj1 oncogene) or (oncogene adj1 KRAS2) or (transforming adj1 protein adj1 p21) or (v adj1 Ki adj1 ras2 adj1 Kirsten adj1 rat adj1 sarcoma adj1 2 viral adj1 oncogene adj1 homolog) | 13831 |
| 11 | K-RAS + RNAi | 10 and 1 | 1378 | |
| 12 | K-RAS + RNAi + IPC | 11 and 6 | 1246 (Unique 753) | |
| 13 | VEGF | ((Vascular ADJ1 endothelial ADJ1 growth ADJ1 factor*) OR (Vascular ADJ1 permeability ADJ1 factor*) OR VEGF* OR VPF OR MGC70609) | 42990 | |
| 14 | VEGF + RNAi + IPC | 1 and 13 and 6 | 5122 (Unique 2718) | |
| 15 | N-RAS | Full Patent Spec | NRAS or (neuroblastoma adj1 RAS adj1 viral adj1 oncogene adj1 homolog) or ALPS4 or N-ras or NRAS1 or NRAS or HRAS1 or (C adj1 BAS adj1 HAS) or P21RAS or RASH1 | 5333 |
| 16 | N-RAS + RNAi | 1 and 15 | 674 | |
| 17 | N-RAS + RNAi + IPC | 16 and 6 | 610 (Unique 375) |
Sample Records
| S.No. | Patent/Publication No. | Title | Abstract | Sequence ID/Info |
| 1 | US7556944B2 Source | Methods and compositions for use in preparing siRNAs. | Methods and compositions for producing siRNAs, e.g., in the form of a d-siRNA composition, from dsRNAs are provided. In the subject methods, a dsRNA is contacted with a composition that includes an activity that cleaves dsRNA into siRNAs, where the composition efficiently cleaves dsRNA into siRNAs. siRNAs produced by the subject methods find use in a variety of applications, particularly in applications where the specific reduction or silencing of a gene is desired. Also provided are kits for use in practicing the subject invention. | |
| 2 | US7374935B2 Source | Human Rgr oncogene and truncated transcripts thereof detected in T cell malignancies, antibodies to the encoded polypeptides and methods of use. | Naturally-occurring variants of human Rgr oncogene protein, in particular, abnormally truncated variants found in T cell malignancies, as well as the human Rgr protein are encompassed by the present invention. Also included are antibodies thereto and nucleic acid molecules encoding human Rgr protein and naturally-occurring variants thereof. The present invention further provides methods for diagnosing and treating T cell malignancies associated with abnormally truncated transcripts of human rgr oncogene and/or abnormal truncation of human Rgr protein. | Seq ID 25, 26, 27,28 |
| 3 | US20070072821A1 Source | Genetic polymorphisms associated with cardiovascular disorders and drug response, methods of detection and uses thereof. | The present invention is based on the discovery of genetic polymorphisms that are associated with cardiovascular disorders, particularly acute coronary events such as myocardial infarction and stroke, and genetic polymorphisms that are associated with responsiveness of an individual having a cardiovascular disorder to treatment of the disorder with statin. In particular, the present invention relates to nucleic acid molecules containing the polymorphisms, variant proteins encoded by such nucleic acid molecules, reagents for detecting the polymorphic nucleic acid molecules and proteins, and methods of using the nucleic acid and proteins as well as methods of using reagents for their detection. | The SNPs of the present invention are also useful for designing RNA interference reagents. |
| 4 | US20070077628A1 Source | Polynucleotides, polypeptides and antibodies and use thereof in treating tsg101-associated diseases | An isolated polynucleotide is provided. The isolated polynucleotide encodes a polypeptide having a sequence of at least 10 and no more than 500 amino acids, wherein said sequence is derived from the amino acid sequences of SEQ ID NO: 2, 4 or 6. Also provided are compositions and methods using same for treating Tsg101 associated diseases. | Seq ID 45 to 48 |
| 5 | US20070093438A1 Source | Use of eukaryotic genes affecting spindle formation or microtubule function during cell division for diagnosis and treatment of proliferative diseases. | The present invention relates to the significant functional role of several C. elegans genes and of their corresponding gene products in spindle formation or microtubule function during cell division that could be identified by means of RNA-mediated interference (RNAi) and to the identification and isolation of functional orthologs of said genes including all biologically functional derivatives thereof The invention further relates to the use of said genes and gene products (including said orthologs) in the development or isolation of anti-proliferative agents, particularly their use in appropriate screening assays, and their use for diagnosis and treatment of proliferative and other diseases. In particular, the invention relates to the use of small interfering RNAs derived from said genes for the treatment of proliferative diseases. | |
| 6 | EP1352061B1 Source | Method for inhibiting the expression of a target gene. | The invention relates to a method for inhibiting the expression of a target gene in a cell, comprising the following steps: introduction of an amount of at least one dual-stranded ribonucleic acid (dsRNA I) which is sufficient to inhibit the expression of the target gene. The dsRNA I has a dual-stranded structure formed by a maximum of 49 successive nucleotide pairs. One strand (as1) or at least one section of the one strand (as1) of the dual-stranded structure is complementary to the sense strand of the target gene. The dsRNA has an overhang on the end (E1) of dsRNA I formed by 1 - 4 nucleotides. | Seq ID of Target gene- MDR1 gene (SEQ ID NO:1-140) |
| 7 | EP1370693B1 Source | Amplified cancer gene WIP1. | There are disclosed methods and compositions for the diagnosis, prevention, and treatment of tumors and cancers in mammals, for example, humans, utilizing the WIP1 gene, which are amplified breast, and/or lung, and/or colon, and/or ovarian, and/or prostate cancer genes. The WIP1 gene, its expressed protein products and antibodies are used diagnostically or as targets for cancer therapy; they are also used to identify compounds and reagents useful in cancer diagnosis, prevention, and therapy. | Target gene - WIP1, Seq ID 1 |
| 8 | US20090175827A1 Source | MiR-16 Regulated Genes and Pathways as targets for therapeutics intervention. | The present invention concerns methods and compositions for identifying genes or genetic pathways modulated by miR-16, using miR-16 to modulate a gene or gene pathway, using this profile in assessing the condition of a patient and/or treating the patient with an appropriate miRNA. | miR-16 uagcagcacguaaauauuggcg SEQ ID NO:1 (MIMAT0000069) |
| 9 | US20090163564A1 Source | Translational Dysfunction Based Therapeutics. | Provided are methods and compositions for inhibiting eukaryotic translation initiation factor eIF4E. Such methods and compositions may be used alone or in conjunction with other therapies, such as gene therapies, for inhibiting cell proliferation and/or treating cancer. | CGGCTTGAAGATGTACTCTAT-3& 8242; (VEGF, SEQ ID NO: 9). |
| 10 | US20090093050A1 Source | DNA Vaccine Enhancement with MHC Class II Activators. | Methods for treating or preventing hyperproliferating diseases, e.g., cancer, are described. A method may comprise administering to a subject in need thereof a therapeutically effective amount of a nucleic acid encoding an MHC class I and/or II activator and optionally a nucleic acid encoding an antigen. | The sequence of DNA encoding the anti-apoptotic protein Bcl2 is SEQ ID NO:77; the amino acid sequence of Bcl2 is SEQ ID NO:78; the PSG5 vector encoding Bcl2, designated PSG5-BCL2, has the sequence SEQ ID NO:79 (with the Bcl2 sequence corresponding to nucleotides 1061 to 1678). |
| 11 | US20090163434A1 Source | MIR-20 Regulated Genes and Pathways as Targets for Therapeutic Intervention. | The present invention concerns methods and compositions for identifying genes or genetic pathways modulated by miR-20a, using miR-20a to modulate a gene or gene pathway, using this profile in assessing the condition of a patient and/or treating the patient with an appropriate miRNA. | miR-20 is 100% identical to SEQ ID NO:1 to SEQ ID NO:269. |